Literature: V. Yurchenko, J. Lukes, X. Xu, D. A. Maslov (2006) An Integrated Morphological and Molecular Approach to a New Species Description in the Trypanosomatidae: the Case of Leptomonas podlipaevi n. sp., a Parasite of Boisea rubrolineata (Hemiptera: Rhopalidae). Journal of Eukaryotic Microbiology, v. 53, n. 2, pp.103-111. pdf
Georgaphic distribution and biotope: The host, western boxelder bug, Boisea rubrolineata Barber (family Rhopalidae, order Hemiptera; formerly named Leptocoris rubrolineatus) is broadly distributed throughout the western United States and Canada. A similar species, Boisea trivittata (Leptocoris trivittatus) is found east of the Rockies. The insects are found on boxelder, maple and ash trees. Axenic cultures (UCR 4-1b1 and UCR 5-1b1) were isolated from the hosts collected on the campus of the University of California, Riverside, in November 2002. Clonal lines 4-5 and 5-10-2, derived from the respective cultures, are deposited in the ATCC as type cultures (PRA-171 and PRA-172)
Previously, Blastocrithidia leptocoridis Wallace and Dyer, 1960 (ATCC 30265) was isolated from B. trivittata. Analysis of SL gene repeats confirmed that B. leptocoridis and L. podlipaevi are different species.
General morphology in host and culture: Cells in the host intestine represented typical promastigotes and, less frequently, shorter elongated cells with no visible flagellum. Cells in culture were represented by similar forms, although the short form was predominant..
Cells form round colonies on the BHI agar. Colonies are composed exclusively from the short, aflagellated type. Flagellated long cells reappear upon cultivation in the liquid medium (BHI with 10 ug/ml hemin).
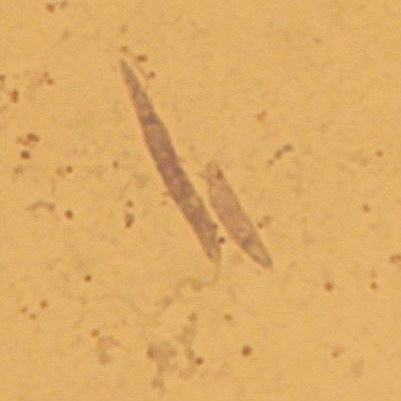
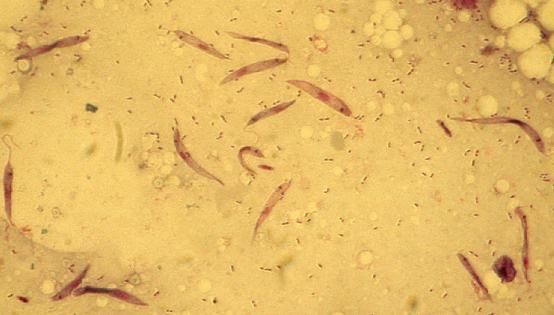
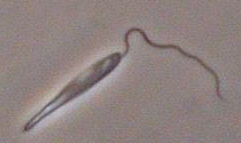

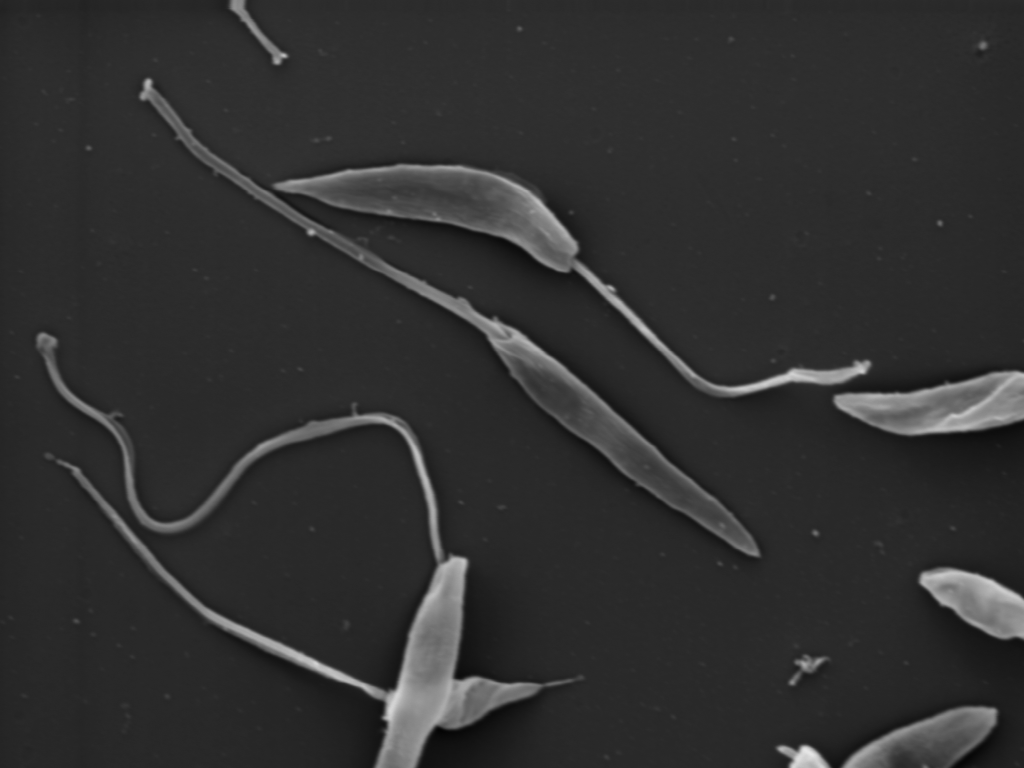
Ultrastructure: The work has been performed in the laboratory of J. Lukes. Details are presented in the reference above.
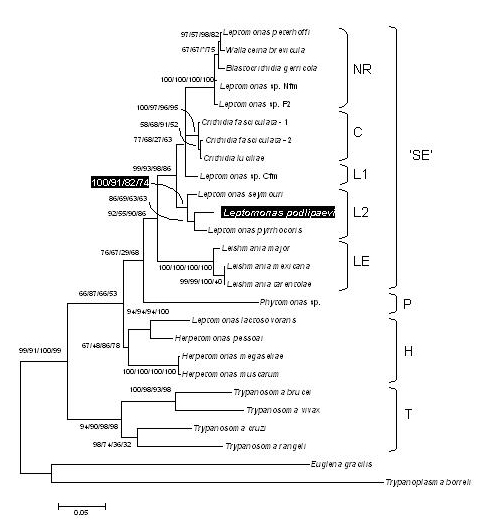
Genotyping and molecular phylogenetic classification. According to the GAPDH phylogenetic analysis, this species is most closely related to Leptomonas seymori and Leptomonas pyrrhocoris. See the publication for details.